Research Overview
|
There
is
increasing evidence in support of a model of cellular biochemistry in
which most proteins exert their biological role through either
transient or relatively stable multi-component macromolecular
complexes. The key to understanding the function of these complexes
lies in their structural investigation by a variety of biophysical
methods. The lab studies molecular details of large protein-nucleic
acid macromolecules using a variety of new NMR techniques as well as
established biophysical approaches. For large complexes, we utilize a
rigid body assembly of individually characterized structures using a
combination of methods: domain orientation through the measurement of
residual dipolar coupling (RDC) by NMR spectroscopy, overall shape
determination by small angle neutron or X-ray scattering (SANS/SAXS),
and incorporating molecular contact details from such techniques as
NMR paramagnetic spin labelling to acquire information on long-range
contacts, as well as in vitro mutational analysis
and other
binding assays. For smaller proteins and domains, standard NMR-based
approaches are used, but with additional insight gained from RDC and
spin label information. Equally important to the lab is the
traditional strength of NMR as a tool to probe the dynamics of
biological samples, the characterization of transient interactions,
and the possibility to look at structures that exhibit a significant
amount of unstructured elements.
Current
projects include:
Nucleic acid recognition
by proteins
Proteins
involved
in RNA processing, especially alternative splicing
Synthetic foldamer
recognition of biomolecules
|
********************************************
NEWS
******************************************
Welcome to Associate Professor Eric Largy who joins our group along
with PhD student Matthieu Ranz!
Congratulations to Sébastien Campagne who will now start his
independent group
at the Institut Européen de Chimie et Biologie (IECB).
Check out his group website https://rna-smart.com/
Previous News Highlights
Current and
past lab members
***********************************
RECENT PUBLICATIONS
*********************************
We are excited to present some of our recent work:
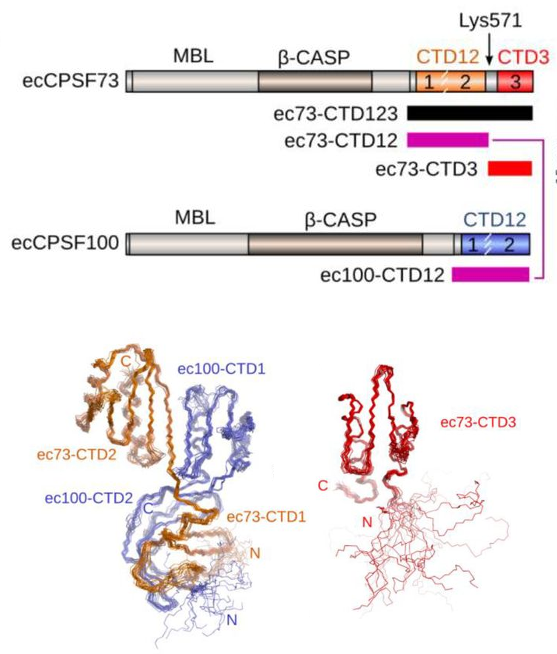 |
Revealing new atomic details in CPSF. Now
published in Open Biology, we report two new NMR structures related to
the
CPSF73 and CPSF100 proteins that are part of the cleavage and
polyadenylation specificity factor - the machinery that is critical in
the processing of the 3'-end of pre-mRNA to generate the polyA tail.
The larger module of the CPSF73-CPSF100 C-terminal heterodimer was
solved by NMR spectroscopy. We had to use some tricks since a complex
of 27 kDa remains a challenging task by NMR. We also solved the
structure of a second module that comprises only a single domain in
CPSF73. We found that this final domain (CTD3) is essential for the
interaction with another component of CPSF known as Symplekin. To be
able to make meaningful alignments, adn also to guide our analysis, we
took advantage of AlphaFold to generate strcuture prediction for the
C-terminal trimer of CPSF73-CPSF100-Symplekin from seven other model
organisms.
|
| Thore,
S., Raoelijaona,
F., Talenton, V., Fribourg, S., Mackereth, C.D. 2023. Molecular details
of the CPSF73-CPSF100 C-terminal heterodimer and interaction with
Symplekin. Open
Biology 13:230221 |
|
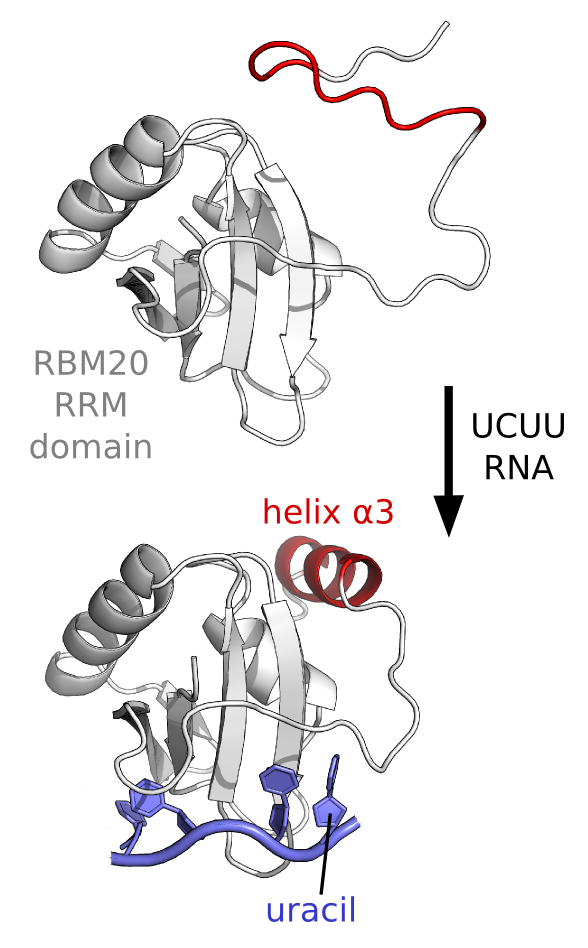
An unusal way to bind RNA. Recent
work from a collaboration with former postoc Santosh Kumar Upadhyay
revealed an uncommon mechanism of RNA-binding by the RNA-Recognition
Module (RRM) domain of a protein factor involved in heart development.
This splicing factor, RBM20, uses the RRM domain to interact with a
UCUU RNA sequence in target mRNA that is important for a healthy heart,
including the mRNA the codes for the titin protein. Mutations in the
RBM20 protein can lead to disease such as dilated cardiomyopathy (DCM).
By using biophysical techniques that include NMR spectroscopy, we have
found that the RRM domain from RBM20 uses a coupled binding-folding
mechanism for selective recognition of the UCUU sequence - in
particular for selectivity towards the last uracil in the RNA motif.
What we found is that an additional helix (helix a3) only forms when
the final uracil is present in the RNA motif, and we also found that
the presence of the final helix is required for high-affinity binding.
|
| Upadhyay,
S.K.,
Mackereth, C.D. 2020. Structural basis of UCUU RNA motif recognition by
splicing factor RBM20. Nucleic
Acids Res. 48:4538-4550. |
All Publications
***************************************************************************************************
|